Our Services
Metagenomics Amplicon Sequencing
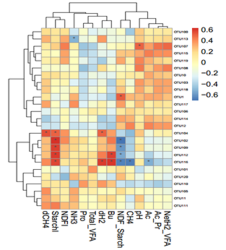
With the use of Illumina NGS technology spectrum to receive prime quality sequencing data sets enabling a comprehensive view into the diversity profile of your microbial community of interest. This platform allows the researcher to retrieve information for bacterial identification and abundance, get evolutionary profiles and structure.
Metagenomic sequencing is culture-independent (useful for uncultured microorganism) and high-throughput analysis. It covers the detection of the microbiota of bacterial, viral or fungal composition in different samples (e.g. human or animal associated, environmental or food samples)
Metagenomic focuses on specific marker genes for phylogenetic identification. For this targeted amplicon approach, the ribosomal small subunit RNA (16S rRNA) gene has emerged as the most widely used marker. Other rRNA genes, the ITS region, COI, or other functional genes can also be used as marker.
Bacterial and Fungal Identification
Our service includes DNA extraction, PCR amplification, DNA purification, and sequencing. The bacterial ID is done by using 16S rRNA gene sequencing which uses PCR with 27F and 1492R primer set to amplify more than 1,300 base pairs. The resulting sequences from the two primers are aligned and BLAST search with NCBI database which gives identification up to genus and species levels.
For fungal ID, 25S rRNA gene or ITS1/ITS2 sequencing is used.
Other services include:
Sanger sequencing
Next generation sequencing
Customised oligonucleotides and fluorescent probes synthesis
Peptides synthesis